2022
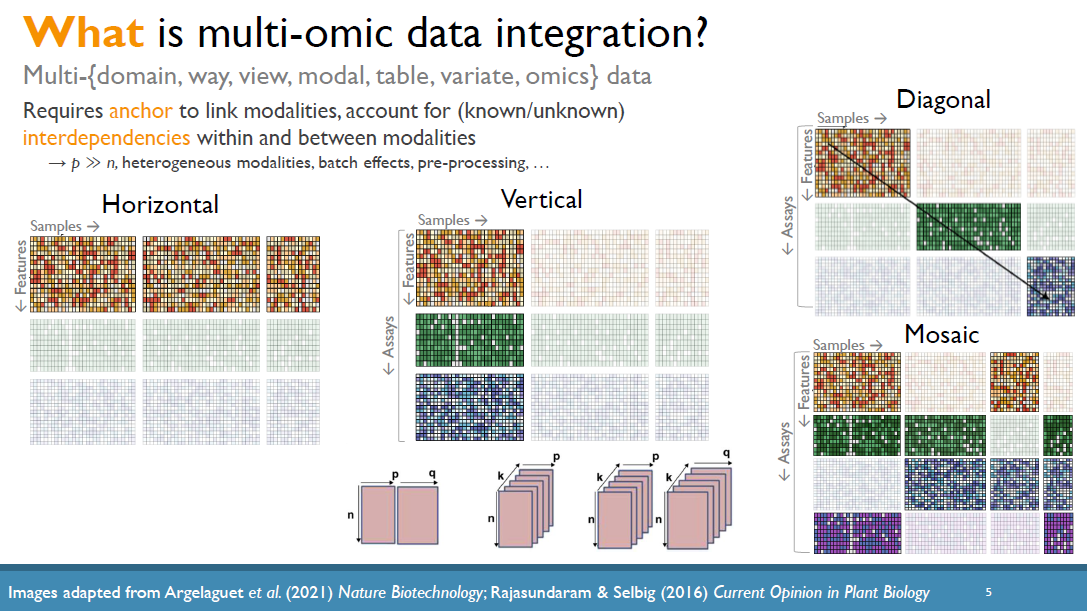
Leveraging multi-omic data for integrative exploratory, predictive, and network analyses
The increased availability and affordability of high-throughput sequencing technologies in recent years have facilitated the use of multi-omic studies, expanding and enriching our understanding of complex systems across hierarchical biological levels. Integrative methods for these heterogeneous and multi-faceted omics data have shown promise for enhancing the interpretability of exploratory analyses, improving predictive power, and contributing to a holistic understanding of systems biology. However, such integrative analyses are accompanied by several major obstacles, including the potentially ambiguous relationships among omic levels, high dimensionality coupled with small sample sizes, technical artefacts due to batch effects, potentially incomplete or missing data… and the occasional difficulty in posing well-defined and answerable research questions of such data. In light of these challenges, in this talk I will discuss a few of our recent methodological contributions to integrate multi-omic data for (1) exploratory analyses, (2) genomic prediction, and (3) network inference, all with a focus on enhanced interpretability and user-friendly software implementations.
(Invited talk) A randomized pairwise likelihood method for complex statistical inferences
Pairwise likelihood methods are commonly used for inference in parametric statistical models in cases where the full likelihood is too complex to be used, such as multivariate count data. Although pairwise likelihood methods represent a useful solution to perform inference for intractable likelihoods, several computational challenges remain, particularly in higher dimensions. To alleviate these issues, we consider a randomized pairwise likelihood approach, where only summands randomly sampled across observations and pairs are used for the estimation. In addition to the usual tradeoff between statistical and computational efficiency, we show that, under a condition on the sampling parameter, this two-way random sampling mechanism allows for the construction of less computationally expensive confidence intervals. The proposed approach, which is implemented in the rpl R package, is illustrated in tandem with copula-based models for multivariate count data in simulations and on a set of transcriptomic data.
2020
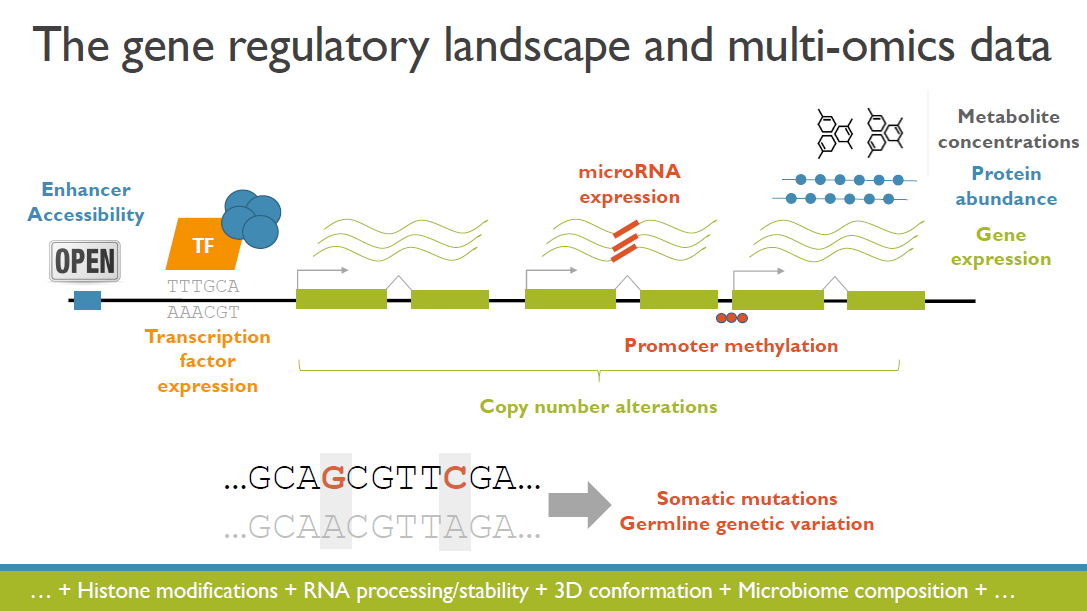
(Invited keynote) Integrative and interactive analyses of multi-omics data
The increased availability and affordability of high-throughput sequencing technologies in recent years has facilitated the use of multi-omic studies to expand and enrich our understanding of complex biological systems. However, defining a holistic and meaningful way to exploit these heterogeneous and multi-faceted ‘omics data can be complicated by several major obstacles. These include the unknown hierarchy and potentially ambiguous relationships among different sources of data, the explosion in data dimension, issues due to batch effects and quality control, potentially incomplete or missing data, limited sample sizes, and the occasional difficulty in posing well-defined and answerable research questions of such data. In light of these challenges, in this talk I will provide an overview of some of our methodological contributions to integrative multi-omic analyses, and I will discuss how the development of interactive tools can be a useful addition to the multi-omic analysis toolbox.
2019
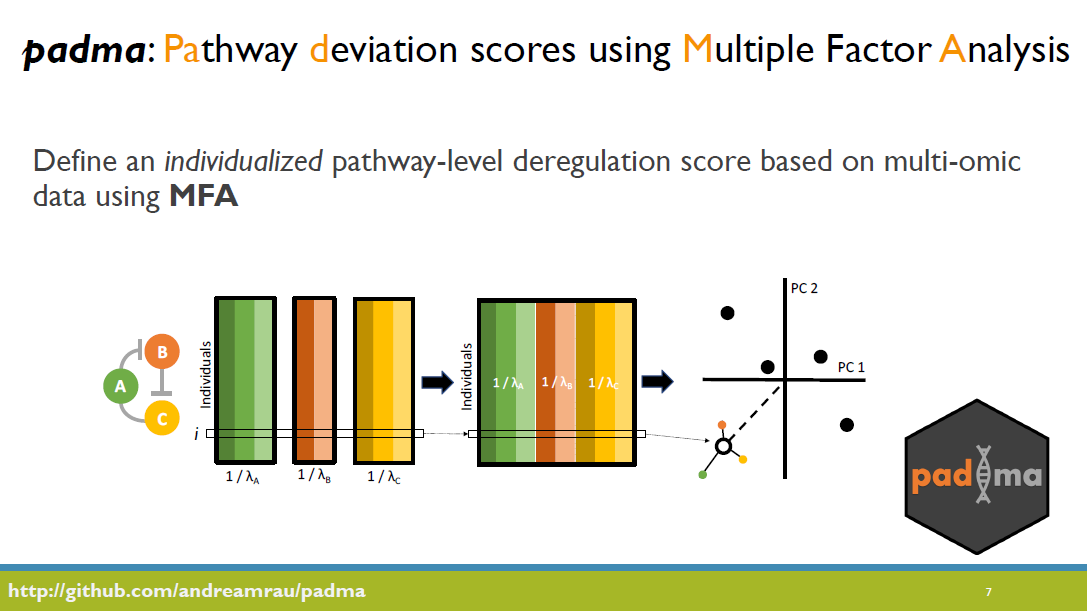
(Invited talk) Individualized multi-omic pathway deviation scores using multiple factor analysis
Malignant progression of normal tissue is typically driven by complex networks of somatic changes, including genetic mutations, copy number aberrations, epigenetic changes, and transcriptional reprogramming. To delineate aberrant multi-omic tumor features that correlate with clinical outcomes, we present a novel pathway-centric tool based on the multiple factor analysis framework called padma. Using a multi-omic consensus representation, padma quantifies and characterizes individualized pathway-specific multi-omic deviations and their underlying drivers, with respect to the sampled population. We demonstrate the utility of padma to correlate patient outcomes with complex genetic, epigenetic, and transcriptomic perturbations in clinically actionable pathways in breast and lung cancer.
- ← Newer
- 2 of 2
- Older →